Creating a ggplot2 plot with a fitted nls model
I have recently helped a colleague to add the curve from a nls model to its ggplot.
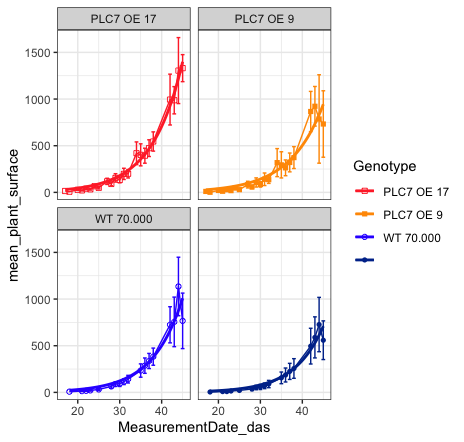
Final result (plot)
As someone mentioned before “let them eat the cake first” before giving the recipe. So here’s the cake / plot!
Input data
The input table (first 10 lines) looks like this:
> head(Plantsize_PLC7_WT, n = 10)
X Plant3.ID Genotype MeasurementDate_das gene Device stress GT ExperimentID projPlantSurfaceArea_mm2 projPlantSurfaceArea_pixels2
1 1 1395478 PLC7 OE 9 17 OE Growscreen 2D 5 control PLC7 790 10.063 8463
2 2 1395478 PLC7 OE 9 20 OE Growscreen 2D 5 control PLC7 790 21.520 18098
3 3 1395478 PLC7 OE 9 22 OE Growscreen 2D 5 control PLC7 790 32.524 27353
4 4 1395478 PLC7 OE 9 24 OE Growscreen 2D 5 control PLC7 790 45.753 38478
5 5 1395478 PLC7 OE 9 27 OE Growscreen 2D 5 control PLC7 790 66.382 55827
6 6 1395478 PLC7 OE 9 29 OE Growscreen 2D 5 control PLC7 790 89.233 75045
7 7 1395478 PLC7 OE 9 31 OE Growscreen 2D 5 control PLC7 790 121.710 102358
8 8 1395478 PLC7 OE 9 34 OE Growscreen 2D 5 control PLC7 790 173.157 145625
9 9 1395478 PLC7 OE 9 35 OE Growscreen 2D 5 control PLC7 790 177.728 149469
10 10 1395478 PLC7 OE 9 42 OE Growscreen 2D 5 control PLC7 790 664.268 558649
Code
And the pièce de résistance, the actual R code:
library("tidyverse")
Plantsize_PLC7_WT <- read.csv("~/Downloads/Plantsize_PLC7_WT.csv")
# part 1 = calculate the mean of the plant surface per genotype
Plantsize_PLC7_WT %>%
group_by(GT, Genotype, MeasurementDate_das, stress) %>%
summarise(
mean_plant_surface = mean(
projPlantSurfaceArea_mm2,
na.rm = TRUE),
sd = sd(
projPlantSurfaceArea_mm2,
na.rm = TRUE),
se = sd / 2^0.5) %>%
# part 2 = ggplot with geom_smooth
ggplot(.,
mapping = aes(color = Genotype,
shape = Genotype,
x = MeasurementDate_das,
y = mean_plant_surface)) +
geom_line() +
geom_point() +
geom_errorbar(aes(ymin = mean_plant_surface - se, ymax = mean_plant_surface + se),) +
theme_bw() +
scale_color_manual(values=c("#FF3333","#FF9900", "#3333FF","#003399")) +
scale_shape_manual(values = c(0, 15, 1, 16)) +
facet_wrap(~ Genotype) +
geom_smooth(method = "nls",
# x is mapped to MeasurementDate_das
# y is mapped to mean_plant_surface
formula = y ~ a * exp(b * x),
se = FALSE, # this is important
method.args = list(start = list(a = 0.1, b = 0.1)))
Written on April 24, 2020